Molecular Pathway Analysis
Molecular Pathway Analysis and the Role of CRISPR in Understanding Cell Biology
Molecular pathway analysis has become a cornerstone of modern biology, enabling researchers to unravel the complex networks of interactions that govern cellular functions. These pathways, comprising proteins, nucleic acids, and small molecules, are central to virtually every aspect of life, from cellular metabolism and growth to differentiation and apoptosis. Understanding these pathways is critical for deciphering the mechanisms underlying health and disease, developing new therapies, and advancing personalized medicine.
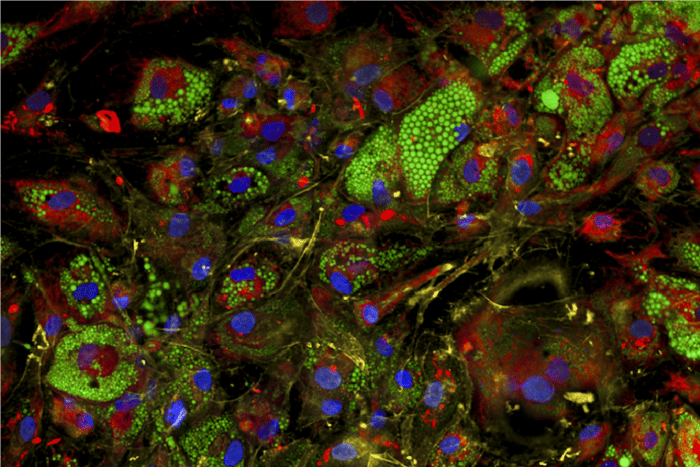
Figure 1. Image-based cellular profiling tools can capture disease-relevant phenotypes. Fat cells, or adipocytes, with stains highlighting nuclei (blue); mitochondria (red); lipid droplets (green); and the actin cytoskeleton, plasma membrane, and Golgi apparatus (yellow). (Credit: Phil Kubitz, Claussnitzer lab, Broad Institute)
Understanding Molecular Pathways
Molecular pathways are essentially maps of biological processes, illustrating how various molecules interact and influence each other to carry out specific cellular functions. These pathways can be broadly categorized into signaling pathways, metabolic pathways, and gene regulatory networks.
- Signaling Pathways: These pathways transmit signals from the cell surface to the interior, influencing cellular responses to external stimuli. Examples include the MAPK/ERK pathway involved in cell division and the PI3K/AKT pathway, which regulates cell survival and metabolism.
- Metabolic Pathways: These are chains of chemical reactions occurring within a cell. They involve the conversion of substrates into products, essential for maintaining cellular function. The glycolysis pathway, which breaks down glucose to produce energy, is a classic example.
- Gene Regulatory Networks: These pathways control the expression of genes, thereby dictating cellular behavior. Transcription factors, non-coding RNAs, and epigenetic modifications are key components of these networks.
Molecular pathway analysis involves the use of computational tools and experimental techniques to map out these pathways, understand their components, and determine how they interact. This analysis is vital for identifying potential drug targets, understanding disease mechanisms, and predicting the effects of genetic mutations.
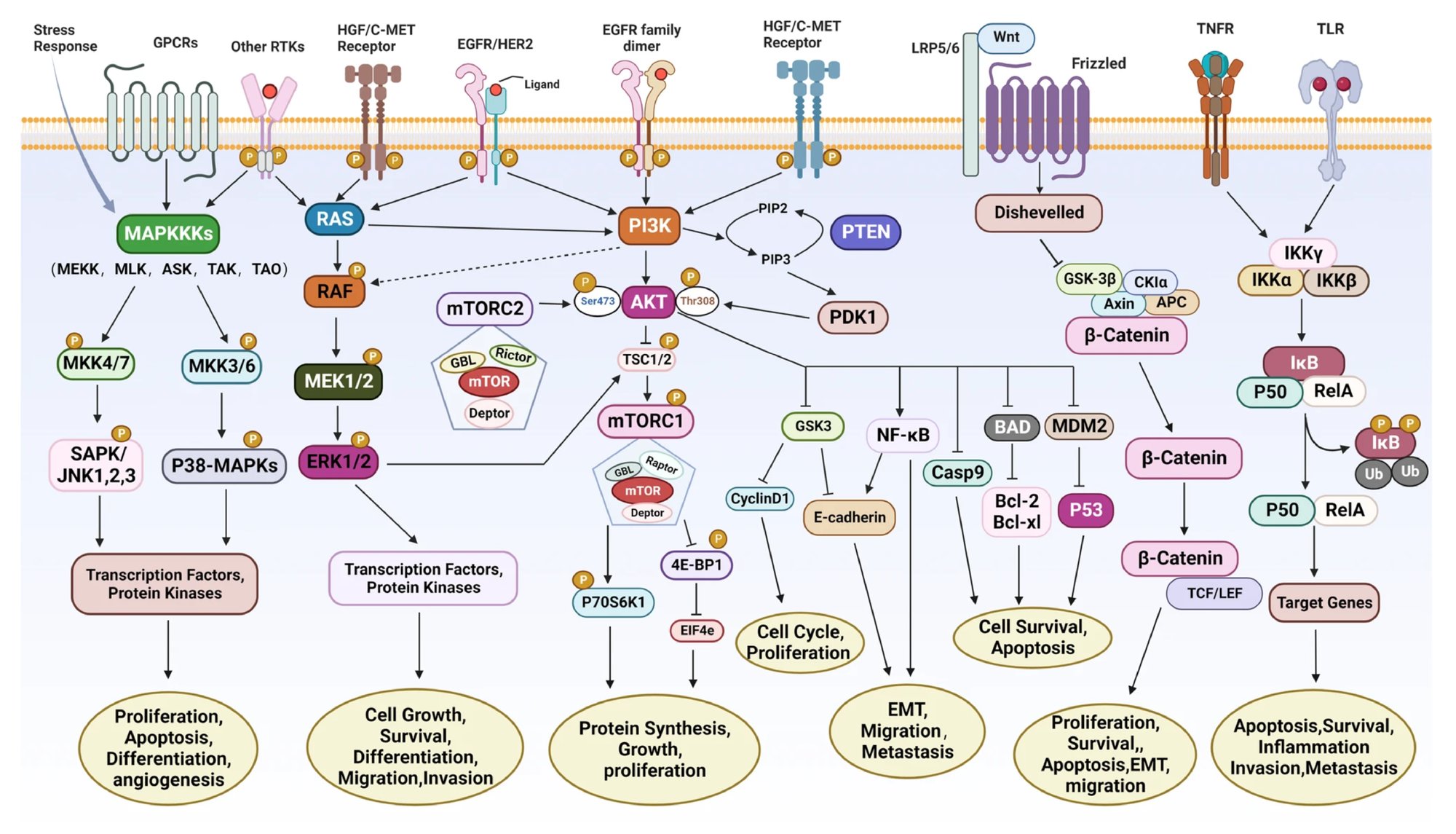
Figure 2. Cancer signaling pathways are complex. (Credit: Signaling pathways and therapeutic interventions in gastric cancer, Nature)
CRISPR in Molecular Pathway Analysis
CRISPR technology has significantly enhanced our ability to study and understand molecular pathways. By enabling precise genetic modifications, CRISPR allows scientists to dissect the roles of individual genes in cellular processes, thereby elucidating the functions of entire pathways.
- Gene Knockout and Knock-in Studies: One of the primary uses of CRISPR in pathway analysis is to create gene knockouts, where specific genes are completely disabled. By observing the effects of these knockouts on cellular function, researchers can identify the role of the gene in a given pathway. For instance, knocking out a gene involved in a signaling pathway can reveal how that gene contributes to signal transduction and cellular responses.
Conversely, CRISPR can also be used to knock in or introduce specific mutations into genes. This is particularly useful for studying the effects of disease-associated mutations on molecular pathways. By mimicking these mutations in cell lines or model organisms, researchers can investigate how they disrupt normal cellular processes and contribute to disease. - Functional Genomic Screens: CRISPR has enabled large-scale functional genomic screens, where thousands of genes can be systematically knocked out or modified to identify their roles in specific pathways. These screens are invaluable for uncovering novel components of pathways, identifying potential drug targets, and understanding genetic interactions.
For example, CRISPR screens have been used to identify genes involved in cancer cell survival, providing insights into the molecular pathways that drive tumor growth and resistance to therapy. These findings can inform the development of new cancer treatments by targeting these pathways. - Epigenome Editing: CRISPR technology has also been adapted for epigenome editing, where it is used to modify the chemical modifications on DNA or histones without altering the underlying genetic code. This is achieved by fusing Cas9 with enzymes that add or remove these modifications. Epigenome editing allows researchers to study the role of epigenetic changes in gene regulation and how they influence molecular pathways.
- Live-Cell Imaging: Recent advancements in CRISPR technology have enabled live-cell imaging of molecular pathways. By tagging specific genes or proteins with fluorescent markers, researchers can visualize the dynamics of these pathways in real-time. This approach has provided new insights into the spatial and temporal regulation of signaling pathways, revealing how they change in response to external stimuli or during disease progression.
Efficient, Precision CRISPR Reagents & Engineered Cells are Streamlining Pathway Analysis
Intelligent Guide Design for Efficient, Consistent Knockouts
EditCo’s innovative approach to smart guide design allows efficient gene disruption generation, eliminating the trial and error associated with common guide design strategies. All our reagents are designed to streamline your research, ensuring more accurate and reliable results.
With intelligent guide design strategy, EditCo is able to simply and effectively knockout genes guaranteed with our Gene Knockout Kits or do high-throughput genomic screening with Arrayed gRNA Libraries, allowing for clear genotype-to-phenotype answers for many genes at scale and quickly.
Simplify Building Gene Knockouts with Edited-to-Order Engineered Cells

We have streamlined the editing step of the knockout and knock-in experimental workflow by completely eliminating the need for scientists to optimize the transfection themselves. Our Engineered Cells family, enabled through our novel, high-throughput CRISPR platform, allows all researchers to affordably access state-of-the-art knockouts and precision knock-ins in pool or clonal formats. EditCo offers a wide range of ATCC-supplied immortalized cell lines, iPS cells, and primary cells – OR – onboard your own cell lines!
For researchers who are studying multiple parts in a pathway and need to systematically knock out each gene, EditCo’s Knockout Cell Pools provide the fastest way to obtain CRISPR knockouts so the researcher can elucidate every gene’s role. Unlike clones, Knockout Cell Pools are a faster, economical, and “bench-free” option so the researcher can obtain and test multiple gene knockouts in parallel, and not miss out on any gene targets. XDel KO pools deliver high editing efficiency, and can be assayed directly to allow quick phenotypic checks without having to wait to generate a clone.
Have more questions? Reach out to us!

